Assemblies
Assembly Name:
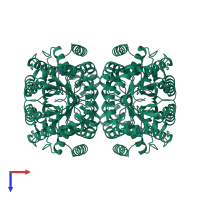
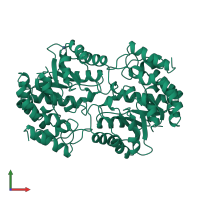
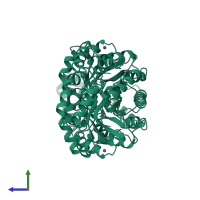
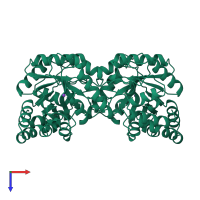
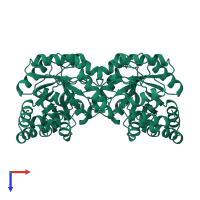
4-hydroxy-tetrahydrodipicolinate synthase
Multimeric state:
homo tetramer
Accessible surface area:
39796.41 Å2
Buried surface area:
8085.41 Å2
Dissociation area:
968.5
Å2
Dissociation energy (ΔGdiss):
-3.77
kcal/mol
Dissociation entropy (TΔSdiss):
14.59
kcal/mol
Symmetry number:
4
PDBe Complex ID:
PDB-CPX-141377
Assembly Name:
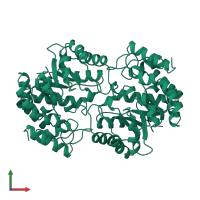
4-hydroxy-tetrahydrodipicolinate synthase
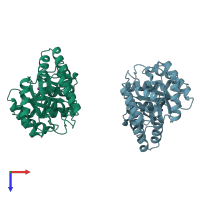
Multimeric state:
homo dimer
Accessible surface area:
20866.7 Å2
Buried surface area:
3074.21 Å2
Dissociation area:
1,298.88
Å2
Dissociation energy (ΔGdiss):
11.13
kcal/mol
Dissociation entropy (TΔSdiss):
13.38
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-141376
Macromolecules
Chains: A, B
Length: 292 amino acids
Theoretical weight: 31.3 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli
UniProt:
Pfam: Dihydrodipicolinate synthetase family
InterPro:
SCOP: Class I aldolase
Length: 292 amino acids
Theoretical weight: 31.3 KDa
Source organism: Escherichia coli
Expression system: Escherichia coli
UniProt:
- Canonical:
 P0A6L2 (Residues: 1-292; Coverage: 100%)
P0A6L2 (Residues: 1-292; Coverage: 100%)
Pfam: Dihydrodipicolinate synthetase family
InterPro:
- DapA-like
- 4-hydroxy-tetrahydrodipicolinate synthase, DapA
- Aldolase-type TIM barrel
- Schiff base-forming aldolase, conserved site
- Schiff base-forming aldolase, active site
SCOP: Class I aldolase