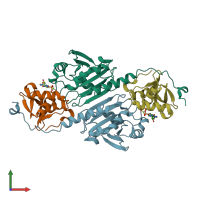
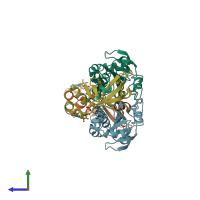
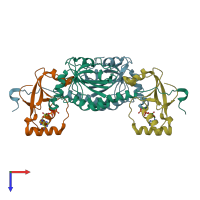
Function and Biology Details
Reactions catalysed:
Thioredoxin + ROOH = thioredoxin disulfide + H(2)O + ROH
Peroxiredoxin-(S-hydroxy-S-oxocysteine) + ATP + 2 R-SH = peroxiredoxin-(S-hydroxycysteine) + ADP + phosphate + R-S-S-R
Biochemical function:
Biological process:
Cellular component:
Structure analysis Details
Assembly composition:
hetero tetramer (preferred)
Assembly name:
Peroxiredoxin-1 and Sulfiredoxin-1 (preferred)
PDBe Complex ID:
PDB-CPX-170402 (preferred)
Entry contents:
2 distinct polypeptide molecules
Macromolecules (2 distinct):