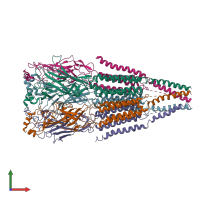
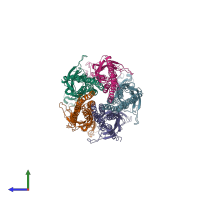
Function and Biology Details
Biochemical function:
Biological process:
Cellular component:
Sequence domains:
- Neurotransmitter-gated ion-channel
- Nicotinic acetylcholine receptor
- Neuronal acetylcholine receptor
- Neurotransmitter-gated ion-channel ligand-binding domain superfamily
- Neurotransmitter-gated ion-channel transmembrane domain
- Neurotransmitter-gated ion-channel transmembrane domain superfamily
- Neurotransmitter-gated ion-channel, conserved site
- Neurotransmitter-gated ion-channel ligand-binding domain
Structure analysis Details
Assembly composition:
hetero pentamer (preferred)
Assembly name:
Acetylcholine receptor, alpha1-beta1-gamma-delta (preferred)
PDBe Complex ID:
PDB-CPX-136096 (preferred)
Entry contents:
4 distinct polypeptide molecules
Macromolecules (4 distinct):