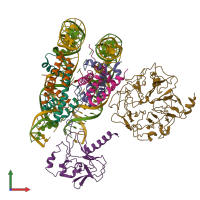
Function and Biology Details
Reaction catalysed:
S-adenosyl-L-methionine + a [histone H4]-L-lysine(20) = S-adenosyl-L-homocysteine + a [histone H4]-N(6)-methyl-L-lysine(20)
Biochemical function:
Biological process:
Cellular component:
Structure analysis Details
Assembly composition:
hetero hexadecamer (preferred)
PDBe Complex ID:
PDB-CPX-118751 (preferred)
Entry contents:
6 distinct polypeptide molecules
2 distinct DNA molecules
2 distinct DNA molecules
Macromolecules (8 distinct):
Ligands and Environments
No bound ligands
No modified residues
Experiments and Validation Details
X-ray source:
APS BEAMLINE 24-ID-E
Spacegroup:
C2221
Expression system: Escherichia coli BL21(DE3)