Function and Biology Details
Structure analysis Details
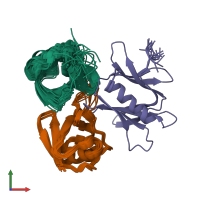
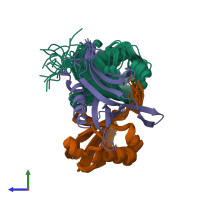
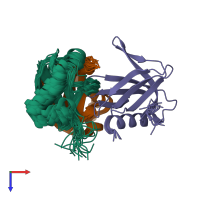
Assembly composition:
hetero trimer (preferred)
Assembly name:
PDBe Complex ID:
PDB-CPX-143236 (preferred)
Entry contents:
3 distinct polypeptide molecules
Macromolecules (3 distinct):
Ligands and Environments
No bound ligands
No modified residues
Experiments and Validation Details
Chemical shift assignment:
12%
Refinement method:
simulated annealing
Expression system: Escherichia coli