EMD-1950
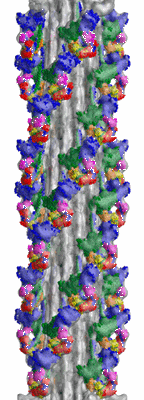
3D-Structure of tarantula myosin filament obtained by cryo-electron microscopy
EMD-1950
Helical reconstruction20.0 Å
 Deposition: 23/08/2011
Deposition: 23/08/2011Map released: 02/09/2011
Last modified: 20/04/2016
Buffer
pH: 7.0
Details: 100mM NaCl,3mM MgCl2,1mM EGTA, 5mM PIPES, 5mM NaH2PO4,1mM NaN3.
Details: 100mM NaCl,3mM MgCl2,1mM EGTA, 5mM PIPES, 5mM NaH2PO4,1mM NaN3.
Staining
Type:
NEGATIVE
Details: A 6 ul aliquot of native purified tarantula thick filaments suspension (Hidalgo et al. 2001) was applied to a 400 mesh grid coated with a holey carbon film that had been rendered hydrophilic by glow discharge in n-amylamine vapor for 3 minutes before use. After allowing the filaments to adsorb to the grid for 30 seconds, the grid was rinsed with the relaxing rinse, then placed in a humidity chamber (aprox. 80% relative humidity). Blotting was performed from one side of the grid till a thin sample film on it using Whatman No 42 filter paper, then the grid was immediately plunged under gravity into liquid ethane cooled by liquid nitrogen. Grids were stored under liquid nitrogen.
Details: A 6 ul aliquot of native purified tarantula thick filaments suspension (Hidalgo et al. 2001) was applied to a 400 mesh grid coated with a holey carbon film that had been rendered hydrophilic by glow discharge in n-amylamine vapor for 3 minutes before use. After allowing the filaments to adsorb to the grid for 30 seconds, the grid was rinsed with the relaxing rinse, then placed in a humidity chamber (aprox. 80% relative humidity). Blotting was performed from one side of the grid till a thin sample film on it using Whatman No 42 filter paper, then the grid was immediately plunged under gravity into liquid ethane cooled by liquid nitrogen. Grids were stored under liquid nitrogen.
Grid
Details: Holey carbon grids 400 mesh
Vitrification
Cryogen name: ETHANE
Chamber humidity: 80%
Chamber temperature: 93 K
Instrument: HOMEMADE PLUNGER
Method: Plunging in a liquid ethane
Details: Vitrification instrument: Home-made plunger. Blotting was performed from one side of the grid till a thin sample film on it using Whatman No 42 filter paper, then the grid was immediately plunged under gravity into liquid ethane cooled by liquid nitrogen. Grids were stored under liquid nitrogen.
Chamber humidity: 80%
Chamber temperature: 93 K
Instrument: HOMEMADE PLUNGER
Method: Plunging in a liquid ethane
Details: Vitrification instrument: Home-made plunger. Blotting was performed from one side of the grid till a thin sample film on it using Whatman No 42 filter paper, then the grid was immediately plunged under gravity into liquid ethane cooled by liquid nitrogen. Grids were stored under liquid nitrogen.
Microscope: FEI/PHILIPS CM120T
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: LAB6
Acceleration voltage: 120 kV
Nominal CS: 2.0 mm
Nominal defocus: 1.95 µm - 1.95 µm
Nominal magnification: 35000.0
Calibrated magnification: 35000.0
Specimen holder model: GATAN LIQUID NITROGEN
Specimen holder details: Eucentric
Details: Holey carbon grids Cryo preserved in Liquid ethane were observed in a Philips CM120 electron microscope under low dose conditions. Only filaments on thin carbon over holes were photographed
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: LAB6
Acceleration voltage: 120 kV
Nominal CS: 2.0 mm
Nominal defocus: 1.95 µm - 1.95 µm
Nominal magnification: 35000.0
Calibrated magnification: 35000.0
Specimen holder model: GATAN LIQUID NITROGEN
Specimen holder details: Eucentric
Details: Holey carbon grids Cryo preserved in Liquid ethane were observed in a Philips CM120 electron microscope under low dose conditions. Only filaments on thin carbon over holes were photographed
Temperature
Minimum: 88
K
Maximum: 90 K
Maximum: 90 K
Image Recording
[1]
Detector category:
FILM
Detector model: KODAK SO-163 FILM
Scanner: OTHER
Sampling interval: 8.47 µm
Number of real images: 1008
Bits per pixel: 14.0
Detector model: KODAK SO-163 FILM
Scanner: OTHER
Sampling interval: 8.47 µm
Number of real images: 1008
Bits per pixel: 14.0
Details: There are 4 helices of myosin heads, rotated 30 degrees, every 145 Angstroms. The filament segments were selected based on visual judgement of good helical order
Final
reconstruction
Resolution: 20.0
Å
(
BY AUTHOR)
Resolution method: FSC 0.5 CUT-OFF
Algorithm: OTHER
Details: Three-dimensional single particle reconstruction was carried out by a modification of the IHRSR method, using SPIDER. Low-dose electron micrographs of 1008 frozen-hydrated thick filaments halves ere digitized at 0.248 nm per pixel using a Nikon Super Coolscan 8000 ED scanner. Filaments were aligned with the bare zone at the top, to ensure correct polarity in subsequent steps. A total of 15,504 segments, each 62 nm long, with an overlap of 55.8 nm, and containing aprox. 40,000 unique pairs of interacting myosin heads went into the reconstruction. As an initial reference model we used the tarantula negatively stained 3D-map, which was axially rotated, axially shifted and also out of plane tilted up to plus-minus12deg. for projection matching, giving a total of 4,095 projections (13 tilted projections plus-minus12deg. every 2deg., 45 reference rotated projections (0-90 degrees, 2deg. rotation angle), and 7 image axial shifts of 2.2 nm. The resulting 3D-map combines about 10,700 out of 15,504 filament segments, a yield of 69 percent of included segments.
Resolution method: FSC 0.5 CUT-OFF
Algorithm: OTHER
Details: Three-dimensional single particle reconstruction was carried out by a modification of the IHRSR method, using SPIDER. Low-dose electron micrographs of 1008 frozen-hydrated thick filaments halves ere digitized at 0.248 nm per pixel using a Nikon Super Coolscan 8000 ED scanner. Filaments were aligned with the bare zone at the top, to ensure correct polarity in subsequent steps. A total of 15,504 segments, each 62 nm long, with an overlap of 55.8 nm, and containing aprox. 40,000 unique pairs of interacting myosin heads went into the reconstruction. As an initial reference model we used the tarantula negatively stained 3D-map, which was axially rotated, axially shifted and also out of plane tilted up to plus-minus12deg. for projection matching, giving a total of 4,095 projections (13 tilted projections plus-minus12deg. every 2deg., 45 reference rotated projections (0-90 degrees, 2deg. rotation angle), and 7 image axial shifts of 2.2 nm. The resulting 3D-map combines about 10,700 out of 15,504 filament segments, a yield of 69 percent of included segments.
⌯ Applied Symmetry
Software
[1]
| Name | Version | Details |
|---|---|---|
| SPIDER | - | - |
Format: CCP4
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: This is a density map of tarantula thick filaments, the initial view is from the Z line perspective, if the map is rotated by 90 degress in x direction, the J motif of the interacting heads features and the backbone subfilaments can be seen clearly
Details: ::::EMDATABANK.org::::EMD-1950::::
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: This is a density map of tarantula thick filaments, the initial view is from the Z line perspective, if the map is rotated by 90 degress in x direction, the J motif of the interacting heads features and the backbone subfilaments can be seen clearly
Details: ::::EMDATABANK.org::::EMD-1950::::
⬡ Geometry
| X | Y | Z | |
|---|---|---|---|
| Dimensions | 250 | 250 | 250 |
| Origin | -124 | -124 | 0 |
| Spacing | 250 | 250 | 250 |
| Voxel size | 2.48 Å | 2.48 Å | 2.482 Å |
Contour list
| Primary | Level | Source |
|---|---|---|
| True | 25.0 | AUTHOR |
