EMD-2328
Structure of the bacterial V-ATPase from Thermus thermophilius
EMD-2328
Electron Crystallography18.0 Å
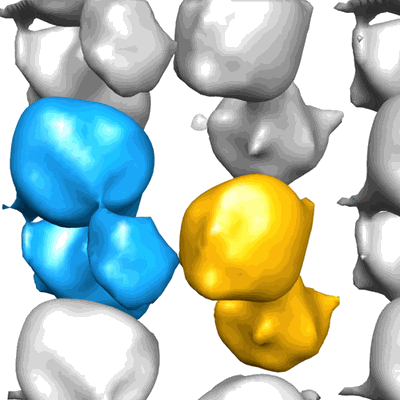 Deposition: 10/03/2013
Deposition: 10/03/2013Map released: 17/04/2013
Last modified: 28/08/2013
Concentration: 1
mg/mL
Details: Crystals grown by dialysis
Details: Crystals grown by dialysis
Staining
Type:
NEGATIVE
Details: A 2.5 ul sample was transferred to a glow-discharged carbon coated copper grid. After 1 min, the drop was blotted with a slice of filter paper, and then the grid was washed with 2.5 ul water to avoid the precipitation caused by mixing phosphate buffer with uranyl acetate. Subsequently, the sample was stained with 2.5 ul of 2% uranyl acetate and air-dried.
Details: A 2.5 ul sample was transferred to a glow-discharged carbon coated copper grid. After 1 min, the drop was blotted with a slice of filter paper, and then the grid was washed with 2.5 ul water to avoid the precipitation caused by mixing phosphate buffer with uranyl acetate. Subsequently, the sample was stained with 2.5 ul of 2% uranyl acetate and air-dried.
Grid
Details: A glow-discharged carbon coated copper grid
Crystal
formation
Details: Crystals grown by dialysis
Microscope: FEI TECNAI F30
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 300 kV
Nominal defocus: 1.4 µm - 2.27 µm
Nominal magnification: 40000.0
Specimen holder model: SIDE ENTRY, EUCENTRIC
Minimum tilt angle: -55
Maximum tilt angle: 55
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 300 kV
Nominal defocus: 1.4 µm - 2.27 µm
Nominal magnification: 40000.0
Specimen holder model: SIDE ENTRY, EUCENTRIC
Minimum tilt angle: -55
Maximum tilt angle: 55
Image Recording
[1]
Detector category:
CCD
Detector model: GENERIC GATAN
Sampling interval: 15 µm
Number of real images: 212
Bits per pixel: 16.0
Details: Every image was recorded by 2Kx2K CCD camera (Gatan)
Detector model: GENERIC GATAN
Sampling interval: 15 µm
Number of real images: 212
Bits per pixel: 16.0
Details: Every image was recorded by 2Kx2K CCD camera (Gatan)
Tilt Series
[1]
| Axis 1 | Axis 2 | |||||
|---|---|---|---|---|---|---|
| Min. | Max. | Inc. | Min. | Max. | Inc. | Rotation |
| -55 ° | 55 ° | - | - | - | - | - |
Format: CCP4
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: Reconstruction of the bacterial V-ATPase
Details: ::::EMDATABANK.org::::EMD-2328::::
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: Reconstruction of the bacterial V-ATPase
Details: ::::EMDATABANK.org::::EMD-2328::::
⬡ Geometry
| X | Y | Z | |
|---|---|---|---|
| Dimensions | 31 | 55 | 73 |
| Origin | -15 | -27 | -36 |
| Spacing | 30 | 54 | 72 |
| Voxel size | 4.296 Å | 4.4 Å | 4.166 Å |
Contour list
| Primary | Level | Source |
|---|---|---|
| True | 0.4 | AUTHOR |
