EMD-2500
Substrate Recruitment Pathways in the Yeast Exosome by Electron Microscopy
EMD-2500
Single-particle25.0 Å
 Deposition: 23/10/2013
Deposition: 23/10/2013Map released: 25/12/2013
Last modified: 12/10/2016
Concentration: 1
mg/mL
Buffer
pH: 8.0
Details: 150mM NaCl, 50mM Tris-HCL,1mM DTT
Details: 150mM NaCl, 50mM Tris-HCL,1mM DTT
Staining
Type:
NEGATIVE
Details: Negatively stained in 2% (w/v) uranyl acetate solution following the standard deep stain procedure on holey-carbon coated EM copper grids covered with a thin layer of continuous carbon
Details: Negatively stained in 2% (w/v) uranyl acetate solution following the standard deep stain procedure on holey-carbon coated EM copper grids covered with a thin layer of continuous carbon
Grid
Details: 300 mesh continuous carbon
Microscope: FEI TECNAI F20
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 200 kV
Specimen holder model: OTHER
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 200 kV
Specimen holder model: OTHER
Image Recording
[1]
Detector category:
CCD
Detector model: GENERIC GATAN (4k x 4k)
Number of real images: 300
Average electron dose per image: 30 e/Å2
Detector model: GENERIC GATAN (4k x 4k)
Number of real images: 300
Average electron dose per image: 30 e/Å2
Final
reconstruction
Resolution: 25.0
Å
(
BY AUTHOR)
Resolution method: FSC 0.5 CUT-OFF
Number of images used: 24913
Resolution method: FSC 0.5 CUT-OFF
Number of images used: 24913
⌯ Applied Symmetry
Point group:
C1
Software
[1]
| Name | Version | Details |
|---|---|---|
| SPIDER | - | - |
⦩ Final angle assignment
Details: SPIDER: theta 90 degrees, phi 359.9 degrees
Format: CCP4
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
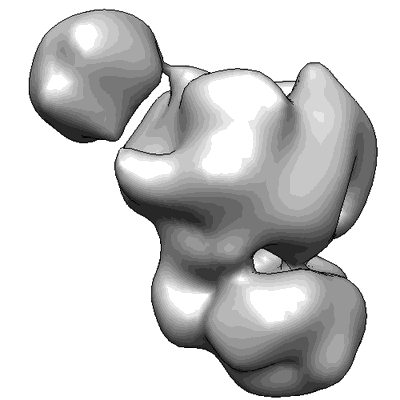
Annotation details: Exosome incubated with streptavidin labeled ssRNA (50 nucleotides in length)
Details: ::::EMDATABANK.org::::EMD-2500::::
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: Exosome incubated with streptavidin labeled ssRNA (50 nucleotides in length)
Details: ::::EMDATABANK.org::::EMD-2500::::
⬡ Geometry
| X | Y | Z | |
|---|---|---|---|
| Dimensions | 90 | 90 | 90 |
| Origin | 0 | 0 | 0 |
| Spacing | 90 | 90 | 90 |
| Voxel size | 3.0 Å | 3.0 Å | 3.0 Å |
Contour list
| Primary | Level | Source |
|---|---|---|
| True | 5.0 | AUTHOR |
