EMD-5617
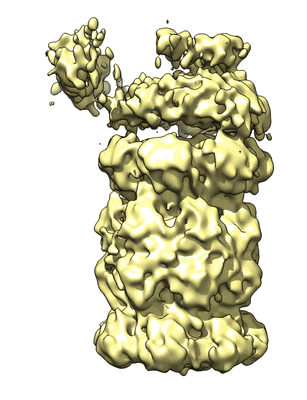
Yeast 20S proteasome in complex with purified yeast 19S base sub complex
EMD-5617
Single-particle9.6 Å
 Deposition: 21/03/2013
Deposition: 21/03/2013Map released: 15/05/2013
Last modified: 05/06/2013
Vitrification
Cryogen name: ETHANE
Chamber humidity: 100%
Chamber temperature: 120 K
Instrument: FEI VITROBOT MARK III
Chamber humidity: 100%
Chamber temperature: 120 K
Instrument: FEI VITROBOT MARK III
Microscope: FEI TECNAI F20
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 200 kV
Nominal CS: 2.0 mm
Nominal defocus: 1.2 µm - 2.1 µm
Nominal magnification: 100000.0
Specimen holder model: GATAN LIQUID NITROGEN
Specimen holder details: CT3500
Minimum tilt angle: 0
Maximum tilt angle: 0
Details: Low-dose procedure
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: FIELD EMISSION GUN
Acceleration voltage: 200 kV
Nominal CS: 2.0 mm
Nominal defocus: 1.2 µm - 2.1 µm
Nominal magnification: 100000.0
Specimen holder model: GATAN LIQUID NITROGEN
Specimen holder details: CT3500
Minimum tilt angle: 0
Maximum tilt angle: 0
Details: Low-dose procedure
Image Recording
[1]
Detector category:
CCD
Detector model: TVIPS TEMCAM-F816 (8k x 8k)
Average electron dose per image: 25 e/Å2
Detector model: TVIPS TEMCAM-F816 (8k x 8k)
Average electron dose per image: 25 e/Å2
Details: FREALIGN
Final
reconstruction
Resolution: 9.6
Å
(
BY AUTHOR)
Resolution method: FSC 0.143 CUT-OFF
Number of images used: 51000
Details:
Resolution method: FSC 0.143 CUT-OFF
Number of images used: 51000
Details:
⌯ Applied Symmetry
Point group:
C1
Software
[1]
| Name | Version | Details |
|---|---|---|
| FREALIGN | - | - |
CTF correction
Details:Each Particle
Format: CCP4
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: 3D density map of yeast 20S proteasome core particle with GST tag in complex with purified yeast 19S base subcomplex
Details: ::::EMDATABANK.org::::EMD-5617::::
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: 3D density map of yeast 20S proteasome core particle with GST tag in complex with purified yeast 19S base subcomplex
Details: ::::EMDATABANK.org::::EMD-5617::::
⬡ Geometry
| X | Y | Z | |
|---|---|---|---|
| Dimensions | 280 | 280 | 280 |
| Origin | 0 | 0 | 0 |
| Spacing | 280 | 280 | 280 |
| Voxel size | 1.47 Å | 1.47 Å | 1.47 Å |
Contour list
| Primary | Level | Source |
|---|---|---|
| True | 0.45 | AUTHOR |
