EMD-6370
3D-Structure of negatively stained Schistosome myosin filament obtained by low-dose electron microscopy
EMD-6370
Helical reconstruction23.0 Å
 Deposition: 02/07/2015
Deposition: 02/07/2015Map released: 07/10/2015
Last modified: 04/11/2015
Buffer
pH: 7.0
Details: 100 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 5 mM PIPES, 1mM NaN3, 5 mM MgATP, 0.01 mM blebbistatin, protease inhibitor cocktail (Sigma P-8465)
Details: 100 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 5 mM PIPES, 1mM NaN3, 5 mM MgATP, 0.01 mM blebbistatin, protease inhibitor cocktail (Sigma P-8465)
Staining
Type:
NEGATIVE
Details: One drop of filament suspension was placed on grids and negatively stained with 1% uranyl acetate.
Details: One drop of filament suspension was placed on grids and negatively stained with 1% uranyl acetate.
Grid
Details: 400-mesh holey carbon grids. Specimens were imaged on thin carbon extending over the holes.
Microscope: FEI/PHILIPS CM120T
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: LAB6
Acceleration voltage: 80 kV
Nominal CS: 2.0 mm
Nominal defocus: 0.6 µm - 2.4 µm
Nominal magnification: 42000.0
Calibrated magnification: 42000.0
Specimen holder model: SIDE ENTRY, EUCENTRIC
Specimen holder details: Room temperature holder
Alignment procedure: LEGACY (Astigmatism: Objective lens astigmatism was corrected at 240,000 times magnification, Electron beam tilt params: )
Details: 1.5 post-magnification, low-dose conditions
Illumination mode: FLOOD BEAM
Imaging mode: BRIGHT FIELD
Electron source: LAB6
Acceleration voltage: 80 kV
Nominal CS: 2.0 mm
Nominal defocus: 0.6 µm - 2.4 µm
Nominal magnification: 42000.0
Calibrated magnification: 42000.0
Specimen holder model: SIDE ENTRY, EUCENTRIC
Specimen holder details: Room temperature holder
Alignment procedure: LEGACY (Astigmatism: Objective lens astigmatism was corrected at 240,000 times magnification, Electron beam tilt params: )
Details: 1.5 post-magnification, low-dose conditions
Image Recording
[1]
Detector category:
CCD
Detector model: TVIPS TEMCAM-F224 (2k x 2k)
Number of real images: 263
Average electron dose per image: 10 e/Å2
Bits per pixel: 16.0
Details: Images were acquired with a 2K x 2K CCD TVIPS camera model F224HD at 5.7 A/pixel.
Detector model: TVIPS TEMCAM-F224 (2k x 2k)
Number of real images: 263
Average electron dose per image: 10 e/Å2
Bits per pixel: 16.0
Details: Images were acquired with a 2K x 2K CCD TVIPS camera model F224HD at 5.7 A/pixel.
Details: 820 thick filament halves were selected from micrographs and stored in SPIDER format. 131 x 131 pixel segments were cut from these filaments, corresponding to a window of 74.7 nm (~five 14.5 nm-spaced crowns of heads).
Final
reconstruction
Resolution: 23.0
Å
(
BY AUTHOR)
Resolution method: OTHER
Algorithm: OTHER
Details: For each iteration of reconstruction (30 cycles), filament segment projections were compared with different projections of the reference reconstruction as follows: seven 2.3 nm axial shifts, 2 degree intervals of rotation about the filament axis up to 90 degrees, and 2 degree intervals of out-of-plane tilting from -10 degrees to +10 degrees. The total number of projections was 7 x 45 x 11 = 3465. For the final 19 cycles of the reconstruction, we used only the best-ordered 420 filament halves (those in which >30% of the segments were found good enough to be used by the reconstruction script in the back-projection in previous cycles). From ~17,000 segments, ~9,500 (56%) were included in the final reconstruction. This final 3D-reconstruction was the average of the last 19 reconstructions between cycles 12 - 30. Its resolution, according to the 0.5 Fourier Shell Correlation (FSC) criterion, was 2.3 nm.
Resolution method: OTHER
Algorithm: OTHER
Details: For each iteration of reconstruction (30 cycles), filament segment projections were compared with different projections of the reference reconstruction as follows: seven 2.3 nm axial shifts, 2 degree intervals of rotation about the filament axis up to 90 degrees, and 2 degree intervals of out-of-plane tilting from -10 degrees to +10 degrees. The total number of projections was 7 x 45 x 11 = 3465. For the final 19 cycles of the reconstruction, we used only the best-ordered 420 filament halves (those in which >30% of the segments were found good enough to be used by the reconstruction script in the back-projection in previous cycles). From ~17,000 segments, ~9,500 (56%) were included in the final reconstruction. This final 3D-reconstruction was the average of the last 19 reconstructions between cycles 12 - 30. Its resolution, according to the 0.5 Fourier Shell Correlation (FSC) criterion, was 2.3 nm.
⌯ Applied Symmetry
Software
[1]
| Name | Version | Details |
|---|---|---|
| SPIDER, EMAN2 | - | - |
Format: CCP4
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
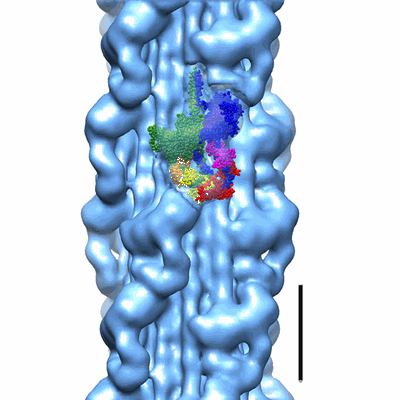
Annotation details: Map of Schistosome thick filaments. Initial view is from the Z-line perspective. If the map is rotated by 90 degrees in x direction, the J motif of the interacting heads is featured and the backbone subfilaments can be seen clearly.
Details: ::::EMDATABANK.org::::EMD-6370::::
Data type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Annotation details: Map of Schistosome thick filaments. Initial view is from the Z-line perspective. If the map is rotated by 90 degrees in x direction, the J motif of the interacting heads is featured and the backbone subfilaments can be seen clearly.
Details: ::::EMDATABANK.org::::EMD-6370::::
⬡ Geometry
| X | Y | Z | |
|---|---|---|---|
| Dimensions | 131 | 131 | 131 |
| Origin | -65 | -65 | 0 |
| Spacing | 131 | 131 | 131 |
| Voxel size | 5.7 Å | 5.7 Å | 5.7 Å |
Contour list
| Primary | Level | Source |
|---|---|---|
| True | 6.0 | AUTHOR |
