Function and Biology Details
Biochemical function:
- not assigned
Biological process:
- not assigned
Cellular component:
- not assigned
Structure analysis Details
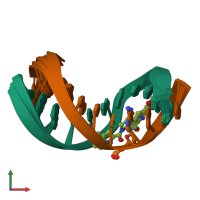
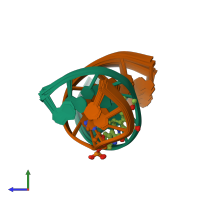
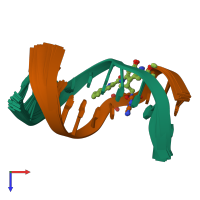
Assembly composition:
hetero dimer (preferred)
Assembly name:
DNA (preferred)
PDBe Complex ID:
PDB-CPX-113710 (preferred)
Entry contents:
2 distinct DNA molecules
Macromolecules (2 distinct):
Ligands and Environments
1 bound ligand:
No modified residues
Experiments and Validation Details
Refinement method:
NAB TO GENERATE A FAMILY OF CONFOMRERS THAT SAMPLE A WIDE RANGE OF
CONFORMATIONAL SPACE. RESTRAINED MOLECULAR DYNAMICS TO GENERATE A RANGE OF
STARTING STRUCTURES FOR DSI. PAIRS OF DNA AND DSI STRUCTURES WERE THEN DOCKED
AND ITERATIVELY REFINED BY A SERIES OF RESTRAINED MOLECULAR DYNAMICS
CALCULATIONS.
Expression system: Not provided