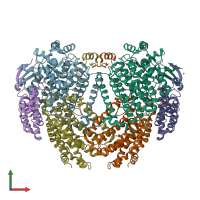
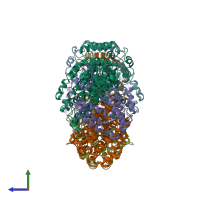
Function and Biology Details
Reaction catalysed:
Methane + NAD(P)H + O(2) = methanol + NAD(P)(+) + H(2)O
Biochemical function:
Biological process:
Cellular component:
- not assigned
Sequence domains:
- Methane monooxygenase, gamma chain
- Methane/phenol monooxygenase, hydroxylase component
- Propane/methane/phenol/toluene hydroxylase
- Ferritin-like superfamily
- Ribonucleotide reductase-like
- Methane monooxygenase, gamma chain, domain 2
- Methane monooxygenase, gamma chain, domain 1
- Methane monooxygenase, gamma chain superfamily
Structure domains:
Structure analysis Details
Assembly composition:
hetero hexamer (preferred)
Assembly name:
Methane monooxygenase A (preferred)
PDBe Complex ID:
PDB-CPX-146024 (preferred)
Entry contents:
3 distinct polypeptide molecules
Macromolecules (3 distinct):