X-ray diffraction
1.96Å resolution
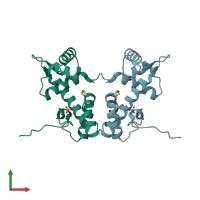
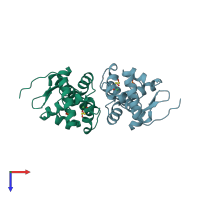
Crystal structure of the nickel-activated two-domain iron-dependent regulator (IdeR)
Released:
DOI:
10.2210/pdb2isy/pdb
Source organism:
Mycobacterium tuberculosis