Function and Biology Details
Reactions catalysed:
Selective hydrolysis of -Xaa-Xaa-|-Yaa- bonds in which each of the Xaa can be either Arg or Lys and Yaa can be either Ser or Ala.
NTP + H(2)O = NDP + phosphate
ATP + H(2)O = ADP + phosphate
Sequence domains:
- P-loop containing nucleoside triphosphate hydrolase
- Helicase superfamily 1/2, ATP-binding domain
- Non-structural protein NS3, peptidase S7, flavivirus
- Helicase, C-terminal domain-like
- Non-structural protein NS3-like, DEAD box helicase, flavivirus
- Peptidase S1, PA clan
- Peptidase S1, PA clan, chymotrypsin-like fold
Structure analysis Details
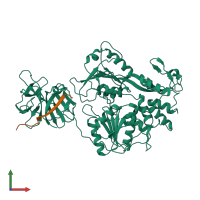
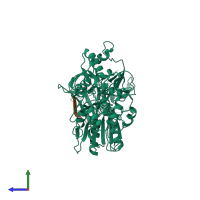
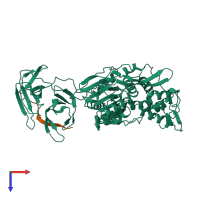
Assembly composition:
hetero dimer (preferred)
Assembly name:
PDBe Complex ID:
PDB-CPX-174050 (preferred)
Entry contents:
2 distinct polypeptide molecules
Macromolecules (2 distinct):
Ligands and Environments
No bound ligands
No modified residues
Experiments and Validation Details
X-ray source:
ESRF BEAMLINE ID23-1
Spacegroup:
P21
Expression system: Escherichia coli BL21