Function and Biology Details
Reactions catalysed:
(1E,2Z)-3-hydroxy-5,9,17-trioxo-4,5:9,10-disecoandrosta-1(10),2-dien-4-oate + H(2)O = 3-((3aS,4S,7aS)-7a-methyl-1,5-dioxo-octahydro-1H-inden-4-yl)propanoate + (2Z,4Z)-2-hydroxyhexa-2,4-dienoate
2,6-dioxo-6-phenylhexa-3-enoate + H(2)O = benzoate + 2-oxopent-4-enoate
Biochemical function:
Biological process:
Cellular component:
Structure analysis Details
Assembly composition:
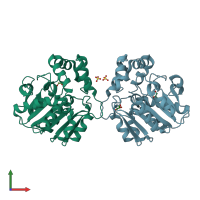
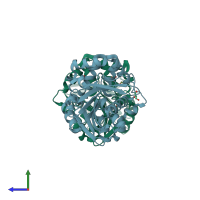
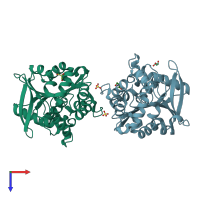
homo tetramer (preferred)
Assembly name:
PDBe Complex ID:
PDB-CPX-161861 (preferred)
Entry contents:
1 distinct polypeptide molecule
Macromolecule: