X-ray diffraction
2.05Å resolution
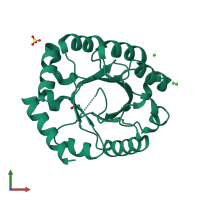
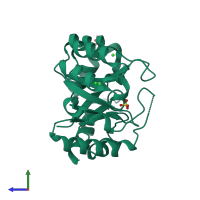
2.05 Angstrom Resolution Crystal Structure of D-ribulose-phosphate 3-epimerase from Francisella tularensis.
Released:
DOI:
10.2210/pdb3inp/pdb
Source organism:
Francisella tularensis subsp. tularensis
Entry authors:
Minasov G, Shuvalova L, Dubrovska I, Winsor J, Scott P, Anderson WF, Center for Structural Genomics of Infectious Diseases (CSGID)