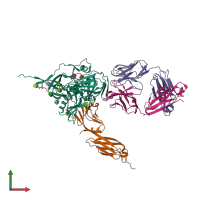
Function and Biology Details
Sequence domains:
- Immunoglobulin-like domain superfamily
- Immunoglobulin-like fold
- Immunoglobulin V-set domain
- Immunoglobulin subtype
- Immunoglobulin-like domain
- Immunoglobulin C1-set
- Human immunodeficiency virus 1, envelope glycoprotein Gp120
- Immunoglobulin/major histocompatibility complex, conserved site
4 more domains
Structure analysis Details
Assembly composition:
hetero tetramer (preferred)
Assembly name:
PDBe Complex ID:
PDB-CPX-208498 (preferred)
Entry contents:
4 distinct polypeptide molecules
Macromolecules (5 distinct):