Function and Biology Details
Reactions catalysed:
Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1)
3'-end directed exonucleolytic cleavage of viral RNA-DNA hybrid
Endohydrolysis of RNA in RNA/DNA hybrids. Three different cleavage modes: 1. sequence-specific internal cleavage of RNA. Human immunodeficiency virus type 1 and Moloney murine leukemia virus enzymes prefer to cleave the RNA strand one nucleotide away from the RNA-DNA junction. 2. RNA 5'-end directed cleavage 13-19 nucleotides from the RNA end. 3. DNA 3'-end directed cleavage 15-20 nucleotides away from the primer terminus.
Specific for a P1 residue that is hydrophobic, and P1' variable, but often Pro.
Biochemical function:
Biological process:
Cellular component:
- not assigned
Structure analysis Details
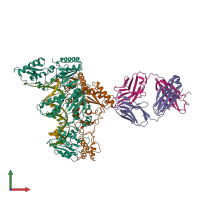
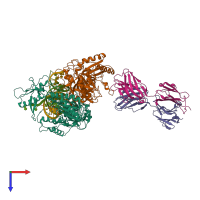
Assembly composition:
hetero hexamer (preferred)
Assembly name:
PDBe Complex ID:
PDB-CPX-207785 (preferred)
Entry contents:
4 distinct polypeptide molecules
2 distinct DNA molecules
2 distinct DNA molecules
Macromolecules (6 distinct):