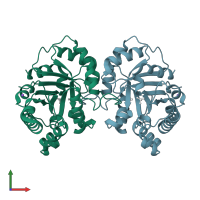
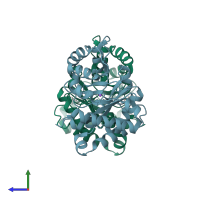
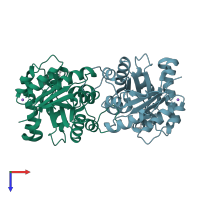
X-ray diffraction
1.55Å resolution
Structure of Triosephosphate Isomerase from Cryptosporidium Parvum at 1.55A Resolution
Released:
DOI:
10.2210/pdb3krs/pdb
Source organism:
Cryptosporidium parvum Iowa II
Primary publication: