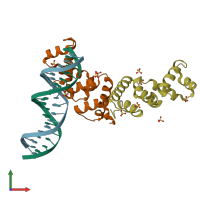
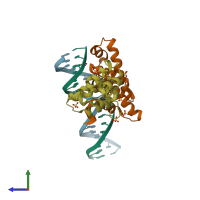Function and Biology Details
Structure analysis Details
Assembly composition:
hetero tetramer (preferred)
Assembly name:
Protein SopB and DNA (preferred)
PDBe Complex ID:
PDB-CPX-116575 (preferred)
Entry contents:
1 distinct polypeptide molecule
1 distinct DNA molecule
1 distinct DNA molecule
Macromolecules (2 distinct):