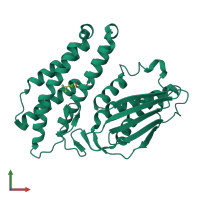
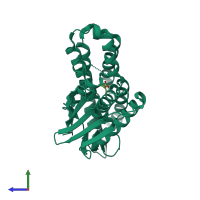
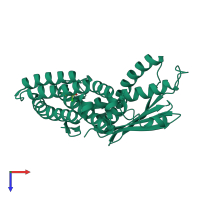
Function and Biology Details
Reactions catalysed:
ATP + a protein = ADP + a phosphoprotein
ATP + [3-methyl-2-oxobutanoate dehydrogenase (acetyl-transferring)] = ADP + [3-methyl-2-oxobutanoate dehydrogenase (acetyl-transferring)] phosphate
Biochemical function:
Biological process:
Cellular component:
Sequence domains:
- Signal transduction histidine kinase-related protein, C-terminal
- Histidine kinase/HSP90-like ATPase
- Histidine kinase/HSP90-like ATPase superfamily
- Alpha-ketoacid/pyruvate dehydrogenase kinase, N-terminal domain superfamily
- Histidine kinase domain
- Branched-chain alpha-ketoacid dehydrogenase kinase/Pyruvate dehydrogenase kinase, N-terminal
- PDK/BCKDK protein kinase
Structure analysis Details
Assemblies composition:
Assembly name:
PDBe Complex ID:
PDB-CPX-169495 (preferred)
Entry contents:
1 distinct polypeptide molecule
Macromolecule: