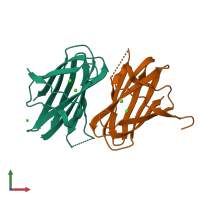Function and Biology Details
Reaction catalysed:
Digestion of native collagen in the triple helical region at -|-Gly bonds. With synthetic peptides, a preference is shown for Gly at P3 and P1', Pro and Ala at P2 and P2', and hydroxyproline, Ala or Arg at P3'.
Biochemical function:
- not assigned
Biological process:
- not assigned
Cellular component:
- not assigned
Sequence domain:
Structure analysis Details
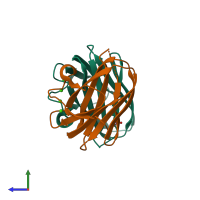
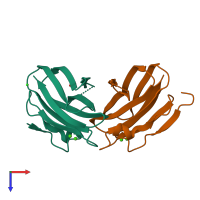
Assembly composition:
hetero dimer (preferred)
Assembly name:
Collagenase ColG (preferred)
PDBe Complex ID:
PDB-CPX-194956 (preferred)
Entry contents:
2 distinct polypeptide molecules
Macromolecules (2 distinct):