Function and Biology Details
Reaction catalysed:
Release of an N-terminal dipeptide, Xaa-Yaa-|-Zaa-, from a polypeptide, preferentially when Yaa is Pro, provided Zaa is neither Pro nor hydroxyproline.
Biochemical function:
Biological process:
Cellular component:
Sequence domains:
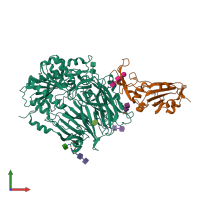
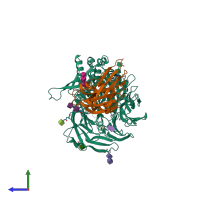
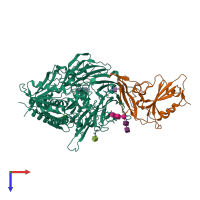
- Spike (S) protein S1 subunit, receptor-binding domain, betacoronavirus
- Alpha/Beta hydrolase fold
- Spike S1 subunit, receptor binding domain superfamily, betacoronavirus
- Spike (S) protein S1 subunit, receptor-binding domain, MERS-CoV
- Peptidase S9, serine active site
- Peptidase S9, prolyl oligopeptidase, catalytic domain
- Dipeptidylpeptidase IV, N-terminal domain
Structure analysis Details
Assembly composition:
hetero tetramer (preferred)
Assembly name:
Spike glycoprotein and Dipeptidyl peptidase 4 (preferred)
PDBe Complex ID:
PDB-CPX-125456 (preferred)
Entry contents:
2 distinct polypeptide molecules
Macromolecules (4 distinct):