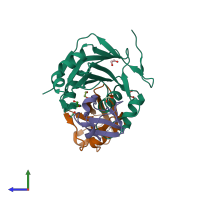X-ray diffraction
2.3Å resolution
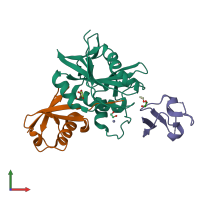
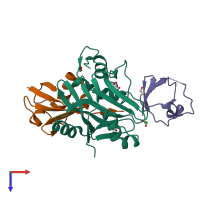
The crystal structure of the DUB domain of AMSH orthologue, Sst2 from S. pombe, in complex with lysine 63-linked diubiquitin
Released:
DOI:
10.2210/pdb4nql/pdb
Source organisms:
Primary publication: