Function and Biology Details
Reactions catalysed:
S-adenosyl-L-methionine + adenine in DNA = S-adenosyl-L-homocysteine + N-6-methyladenine in DNA
Endonucleolytic cleavage of DNA to give specific double-stranded fragments with terminal 5'-phosphates
Biochemical function:
Biological process:
Cellular component:
- not assigned
Sequence domains:
- N4/N6-methyltransferase, Type III restriction-modification enzyme EcoPI Mod subunit-like
- P-loop containing nucleoside triphosphate hydrolase
- S-adenosyl-L-methionine-dependent methyltransferase superfamily
- Type III R-M EcoP15I, C-terminal
- DNA methylase, N-6 adenine-specific, conserved site
- DNA methylase N-4/N-6
- Type III restriction enzyme, C-terminal endonuclease domain
- Helicase/UvrB, N-terminal
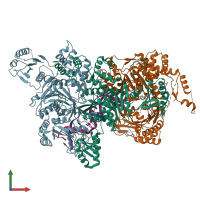
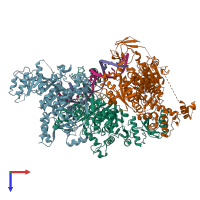
Structure analysis Details
Assembly composition:
hetero pentamer (preferred)
Assembly name:
PDBe Complex ID:
PDB-CPX-118371 (preferred)
Entry contents:
2 distinct polypeptide molecules
2 distinct DNA molecules
2 distinct DNA molecules
Macromolecules (4 distinct):