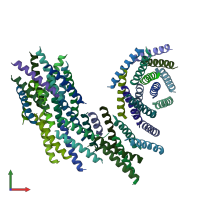
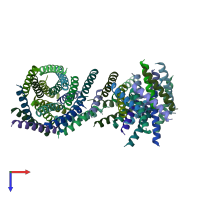
Function and Biology Details
Reaction catalysed:
Release of an N-terminal amino acid, preferably a neutral or hydrophobic one, from a polypeptide. Aminoacyl-arylamides are poor substrates.
Biochemical function:
- not assigned
Biological process:
- not assigned
Cellular component:
- not assigned
Structure analysis Details
Assembly composition:
homo trimer (preferred)
Assembly name:
Vacuolar aminopeptidase 1 (preferred)
PDBe Complex ID:
PDB-CPX-147138 (preferred)
Entry contents:
1 distinct polypeptide molecule
Macromolecule:
Ligands and Environments
No bound ligands
No modified residues
Experiments and Validation Details
X-ray source:
PHOTON FACTORY BEAMLINE AR-NE3A
Spacegroup:
R32
Expression system: Escherichia coli