Function and Biology Details
Reaction catalysed:
All bonds known to be hydrolyzed by this endopeptidase have arginine in P1 and an acidic residue in P4. P6 is often occupied by an acidic residue or by a hydroxy-amino-acid residue, the phosphorylation of which enhances cleavage.
Biochemical function:
Biological process:
Cellular component:
Structure analysis Details
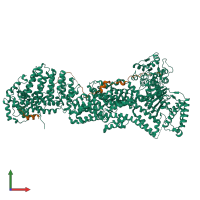
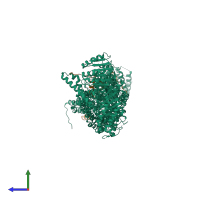
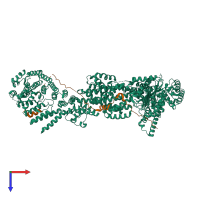
Assembly composition:
hetero dimer (preferred)
Assembly name:
Separase-Securin complex (preferred)
PDBe Complex ID:
PDB-CPX-154072 (preferred)
Entry contents:
2 distinct polypeptide molecules
Macromolecules (2 distinct):
Ligands and Environments
No bound ligands
No modified residues
Experiments and Validation Details
X-ray source:
APS BEAMLINE 24-ID-C
Spacegroup:
P3221
Expression system: Trichoplusia ni