Function and Biology Details
Biochemical function:
Biological process:
Cellular component:
Sequence domains:
- ATP synthase, F0 complex, subunit C
- ATP synthase, F0 complex, subunit C, DCCD-binding site
- V-ATPase proteolipid subunit C-like domain
- F/V-ATP synthase subunit C superfamily
- F1F0 ATP synthase subunit C superfamily
- ATP synthase, F0 complex, subunit A
- ATP synthase, F0 complex, subunit D, mitochondrial
- ATP synthase subunit K
9 more domains
Structure analysis Details
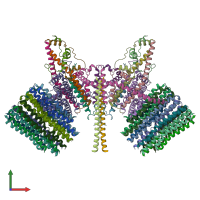
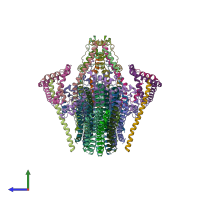
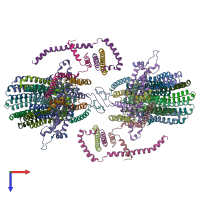
Assembly composition:
hetero 38-mer (preferred)
PDBe Complex ID:
PDB-CPX-133642 (preferred)
Entry contents:
10 distinct polypeptide molecules
Macromolecules (10 distinct):