Function and Biology Details
Biochemical function:
Biological process:
Cellular component:
Sequence domains:
- Ubiquitin/SUMO-activating enzyme E1-like
- THIF-type NAD/FAD binding fold
- ThiF/MoeB/HesA family
- Ubiquitin-activating enzyme
- SUMO-activating enzyme subunit 2, C-terminal domain
- Ubiquitin/SUMO-activating enzyme ubiquitin-like domain
- Ubiquitin-activating enzyme E1, conserved site
- Ubiquitin-activating enzyme E1, Cys active site
4 more domains
Structure analysis Details
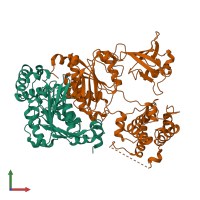
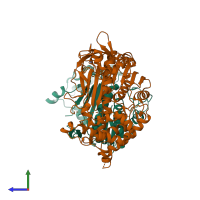
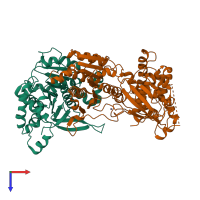
Assembly composition:
hetero dimer (preferred)
Entry contents:
2 distinct polypeptide molecules
Macromolecules (2 distinct):