Function and Biology Details
Biochemical function:
- not assigned
Biological process:
- not assigned
Cellular component:
- not assigned
Structure analysis Details
Assembly composition:
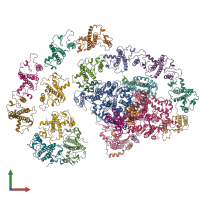
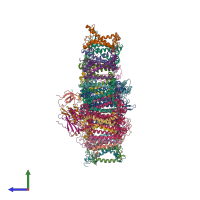
hetero 24-mer (preferred)
Assembly name:
Chlorophyll a/b binding protein 4 and Psad and Psai and Chlorophyll a/b binding protein 2 and Chlorophyll a/b binding protein 6 and Chlorophyll a/b bi... (preferred)
show full assembly name
show full assembly name
Chlorophyll a/b binding protein 4 and Psad and Psai and Chlorophyll a/b binding protein 2 and Chlorophyll a/b binding protein 6 and Chlorophyll a/b binding protein 9 and Psab and Chlorophyll a/b binding protein 8 and Chlorophyll a/b binding protein 10 and Psaa and Psae and Psag and Chlorophyll a/b binding protein 3 and Psal and Chlorophyll a/b binding protein 5 and Psah and Chlorophyll a/b binding protein 12 and Psaf and Chlorophyll a/b binding protein 1 and Psaj and Chlorophyll a/b binding protein 7 and Psac and Psak and Chlorophyll a/b binding protein 11 (preferred)
hide full assembly name
hide full assembly name
PDBe Complex ID:
PDB-CPX-167064 (preferred)
Entry contents:
24 distinct polypeptide molecules
Macromolecules (24 distinct):
Chlorophyll A/B binding protein 1
Chain: A
Molecule details ›
Chain: A
Length: 193 amino acids
Theoretical weight: 16.44 KDa
Source organism: Physcomitrium patens
Length: 193 amino acids
Theoretical weight: 16.44 KDa
Source organism: Physcomitrium patens
Chlorophyll A/B binding protein 5
Chain: B
Molecule details ›
Chain: B
Length: 195 amino acids
Theoretical weight: 16.61 KDa
Source organism: Physcomitrium patens
Length: 195 amino acids
Theoretical weight: 16.61 KDa
Source organism: Physcomitrium patens
Chlorophyll A/B binding protein 3
Chain: C
Molecule details ›
Chain: C
Length: 208 amino acids
Theoretical weight: 17.72 KDa
Source organism: Physcomitrium patens
Length: 208 amino acids
Theoretical weight: 17.72 KDa
Source organism: Physcomitrium patens
Chlorophyll A/B binding protein 7
Chain: D
Molecule details ›
Chain: D
Length: 206 amino acids
Theoretical weight: 17.55 KDa
Source organism: Physcomitrium patens
Length: 206 amino acids
Theoretical weight: 17.55 KDa
Source organism: Physcomitrium patens
Chlorophyll A/B binding protein 4
Chain: E
Molecule details ›
Chain: E
Length: 221 amino acids
Theoretical weight: 18.83 KDa
Source organism: Physcomitrium patens
Length: 221 amino acids
Theoretical weight: 18.83 KDa
Source organism: Physcomitrium patens
Chlorophyll A/B binding protein 9
Chain: F
Molecule details ›
Chain: F
Length: 218 amino acids
Theoretical weight: 18.57 KDa
Source organism: Physcomitrium patens
Length: 218 amino acids
Theoretical weight: 18.57 KDa
Source organism: Physcomitrium patens
Chlorophyll A/B binding protein 8
Chain: G
Molecule details ›
Chain: G
Length: 219 amino acids
Theoretical weight: 18.66 KDa
Source organism: Physcomitrium patens
Length: 219 amino acids
Theoretical weight: 18.66 KDa
Source organism: Physcomitrium patens
Chlorophyll A/B binding protein 2
Chain: H
Molecule details ›
Chain: H
Length: 198 amino acids
Theoretical weight: 16.87 KDa
Source organism: Physcomitrium patens
Length: 198 amino acids
Theoretical weight: 16.87 KDa
Source organism: Physcomitrium patens
Chlorophyll A/B binding protein 6
Chain: I
PsaA
Chain: J
Molecule details ›
Chain: J
Length: 743 amino acids
Theoretical weight: 63.25 KDa
Source organism: Physcomitrium patens
Length: 743 amino acids
Theoretical weight: 63.25 KDa
Source organism: Physcomitrium patens
Chlorophyll A/B binding protein 10
Chain: K
PsaB
Chain: L
Chlorophyll A/B binding protein 11
Chain: M
Molecule details ›
Chain: M
Length: 224 amino acids
Theoretical weight: 19.08 KDa
Source organism: Physcomitrium patens
Length: 224 amino acids
Theoretical weight: 19.08 KDa
Source organism: Physcomitrium patens
PsaC
Chain: N
Chlorophyll A/B binding protein 12
Chain: O
Molecule details ›
Chain: O
Length: 225 amino acids
Theoretical weight: 19.17 KDa
Source organism: Physcomitrium patens
Length: 225 amino acids
Theoretical weight: 19.17 KDa
Source organism: Physcomitrium patens
PsaD
Chain: P
Molecule details ›
Chain: P
Length: 143 amino acids
Theoretical weight: 12.19 KDa
Source organism: Physcomitrium patens
Length: 143 amino acids
Theoretical weight: 12.19 KDa
Source organism: Physcomitrium patens
PsaE
Chain: Q
PsaF
Chain: R
Molecule details ›
Chain: R
Length: 154 amino acids
Theoretical weight: 13.12 KDa
Source organism: Physcomitrium patens
Length: 154 amino acids
Theoretical weight: 13.12 KDa
Source organism: Physcomitrium patens
PsaG
Chain: S
PsaH
Chain: T
PsaI
Chain: U
PsaJ
Chain: V
PsaK
Chain: W
PsaL
Chain: X
Molecule details ›
Chain: X
Length: 157 amino acids
Theoretical weight: 13.38 KDa
Source organism: Physcomitrium patens
Length: 157 amino acids
Theoretical weight: 13.38 KDa
Source organism: Physcomitrium patens