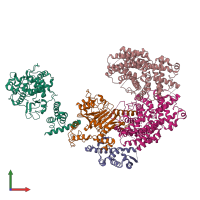
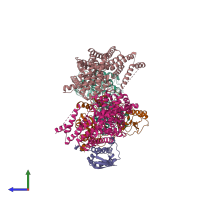
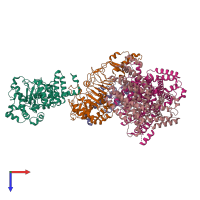
Function and Biology Details
Biochemical function:
Biological process:
Cellular component:
Sequence domains:
- Zn(2)Cys(6) fungal-type DNA-binding domain
- Zn(2)-C6 fungal-type DNA-binding domain superfamily
- S-phase kinase-associated protein 1
- SKP1/BTB/POZ domain superfamily
- Centromere DNA-binding protein complex CBF3 subunit B, C-terminal
- Ndc10, domain 2
- Ndc10, domain 2 superfamily
- SKP1 component, dimerisation
3 more domains
Structure analysis Details
Assembly composition:
hetero pentamer (preferred)
PDBe Complex ID:
PDB-CPX-131699 (preferred)
Entry contents:
4 distinct polypeptide molecules
Macromolecules (4 distinct):