Function and Biology Details
Reaction catalysed:
Ethylene terephthalate + H(2)O = terephthalate + ethylene glycol
Biochemical function:
Biological process:
Cellular component:
Sequence domains:
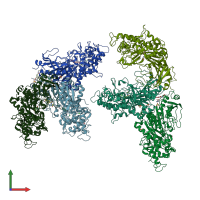
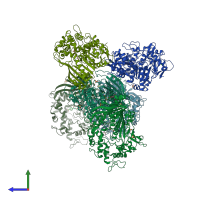
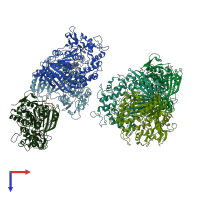
Structure analysis Details
Assembly composition:
monomeric (preferred)
Entry contents:
1 distinct polypeptide molecule
Macromolecule: