Function and Biology Details
Biochemical function:
Biological process:
- not assigned
Cellular component:
Sequence domains:
- Tc toxin complex TcA, C-terminal TcB-binding domain
- Salmonella virulence plasmid 65kDa B protein
- TcA receptor binding domain
- Neuraminidase-like domain
- ABC toxin, N-terminal domain
- Insecticidal toxin complex/plasmid virulence protein
- Rhs repeat-associated core
- Toxin complex C-like repeat
3 more domains
Structure analysis Details
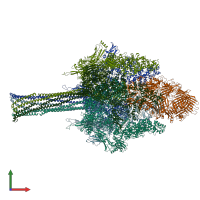
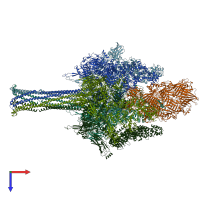
Assembly composition:
hetero hexamer (preferred)
Assembly name:
TcdA1 and TccC3, TcdB2 (preferred)
PDBe Complex ID:
PDB-CPX-184505 (preferred)
Entry contents:
2 distinct polypeptide molecules
Macromolecules (2 distinct):